What Is The Template Strand From The Gel
1.4: PCR and Gel Electrophoresis
- Folio ID
- 73669
Walter Suza; Donald Lee; Marjorie Hanneman; and Deana One thousand. Namuth
Learning Objectives
At the completion of this PCR lesson, learners will exist able to:
- List the 5 chemic components of a PCR reaction and describe their roles.
- List the functions of the 3 temperature cycles which are repeated during a PCR reaction.
- Describe the process of observing results and interpreting results of a PCR experiment.
- List possible uses of PCR in genetic testing and in research.
Overview
The polymerase chain reaction laboratory technique is used in a variety of applications to make copies of a specific DNA sequence. This lesson describes how a PCR reaction works, what information technology accomplishes, and its bones requirements for success. Examples of interpreting results are given. PCR'south strengths, weaknesses, and applications to plant biotechnology are explained.
The Discovery of PCR
In 1983, Kary Mullis was driving forth a Californian mountain road late ane night. As a molecular biologist, Dr. Mullis was imagining a improve way to study DNA. This belatedly-night thinking led to a revolutionary way to make laboratory copies of DNA molecules (Saiki et al. 1985, Mullis 1990). In the decades since, the polymerase chain reaction or PCR, has become the standard method used for detecting specific DNA or RNA sequences. Selling the equipment and reagent kits for PCR is a multi-billion-dollar business concern considering DNA and RNA detection is critical information in many applications.
In Vitro vs. In Vivo Replication
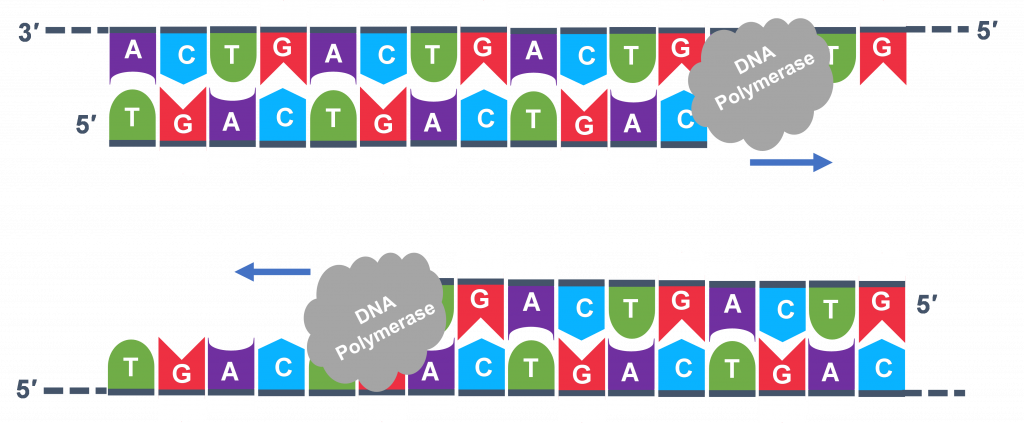
PCR is an In Vitro process; a serial of chemical reactions that happen outside of a living cell. This laboratory technique is modeled later on an In vivo procedure, the living cell'southward natural ability to replicate DNA during normal jail cell cycles (see Lesson on DNA: The Genetic Material). Every living cell makes a indistinguishable copy of each chromosome before the prison cell is set to divide. Figure 1 below illustrates the primal parts of In vivo DNA replication that are the basis for PCR success.
At that place are other enzymes that play an important role in in vivo replication. However, PCR works every bit an in vitro DNA replication process by using only one of these enzymes. Mullis imagined a chemical reagent and a temperature change step in the method that could perform the piece of work of the other 2 enzymes. It should be noted that because Dr. Kerry Mullis had learned most the details of in vivo DNA replication, he could create this scientific discipline changing in vitro method.
Before reading the description of PCR components and processes, watching this video can assistance you visualize the importance of each stride,
Proper noun and Chemical Components of PCR
The name 'Polymerase Concatenation Reaction' represents the nature of the process. 'Polymerase' considering Dna Polymerase III is required for constructing new Dna strands, simply like in a living cell. 'Concatenation Reaction' describes repeating cycles of replication which target a specific segment of a chromosome and use a "copy the copies" progression each bicycle that doubles the amount of Deoxyribonucleic acid copies of a specific segment of DNA present each cycle. In simply 20 cycles of the chain reaction, over one million (ii20) copies of that specific segment of Dna tin can be produced. This is enough DNA to see with your naked centre. The goal of PCR is to make millions of copies of a specific segment of Dna that all originate from a single DNA sample.
The five chemical components that must be added to a exam tube for the PCR reaction to work, include a DNA template, Deoxyribonucleic acid polymerase III enzyme, single stranded DNA primers, nucleotides, and reaction buffer.
- The DNA template is a sample of Dna that contains the target sequence of DNA for copying.
- DNA pol III. There are 2 requirements for a suitable DNA polymerase enzyme for PCR. First, the enzyme must have a good activeness rate around 75°C. Second, the enzyme should be able to withstand temperatures of 95-100°C without denaturing and losing activity.
- 2 primers. Primers are short oligonucleotides of Deoxyribonucleic acid, usually around 20 base of operations pairs in length. Because the purpose of PCR is to amplify a specific section of Dna in the genome, such every bit a known cistron, then primers of specific sequences must be used. The geneticist planning the PCR reaction will design a forward primer to demark to 1 strand and a opposite primer that complements and binds to the other strand. The primer design process to select forward and reverse primers is requiring appropriate genetics thinking and is describe later on in this reading.
- The four dissimilar deoxyribonucleotide triphosphates (dNTPs). Adenine (A), guanine (M), cytosine (C), and thymine (T) are needed to provide the edifice blocks for DNA replication. Dna polymerase will add each complementary base to the new growing Dna strand co-ordinate to the original strand's sequence post-obit normal A-T and C-1000 pairings.
- Finally, a reaction buffer. This creates a stable pH and provides the Mg2+ cofactor needed for DNA pol III activity.
Three Temperature Cycles
A cardinal insight to the success of PCR as an in vitro Dna replication process which generates millions of specific sequence copies was a three-temperature cycle which accomplish three parts of DNA replication: denaturation of the double stranded template, annealing of the primers to the single strands and extension of new strand synthesis by Dna pol 3.
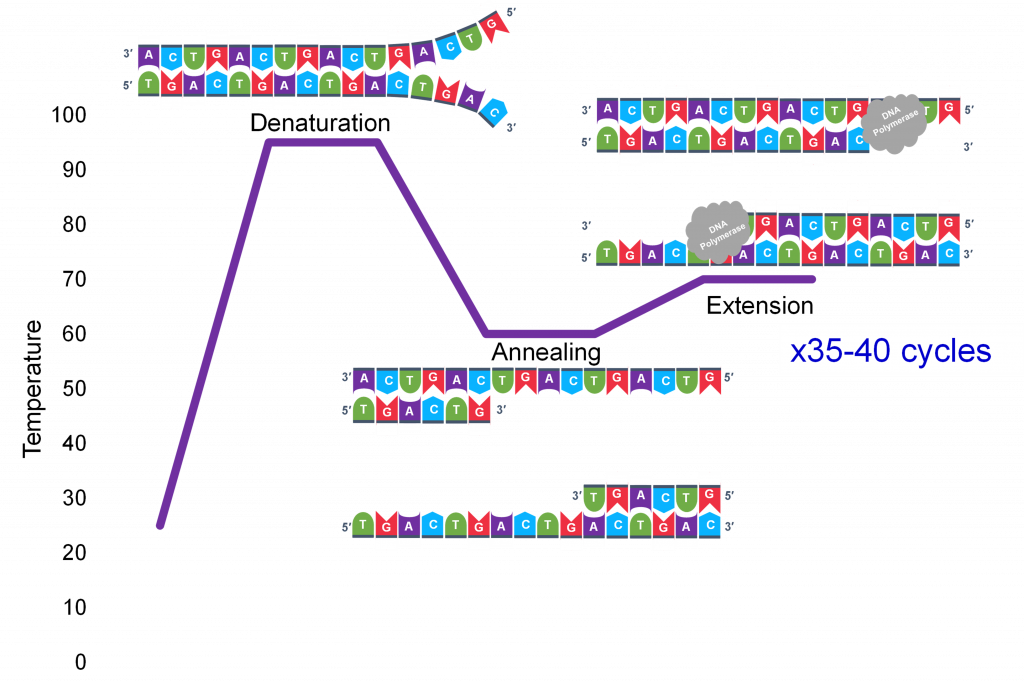
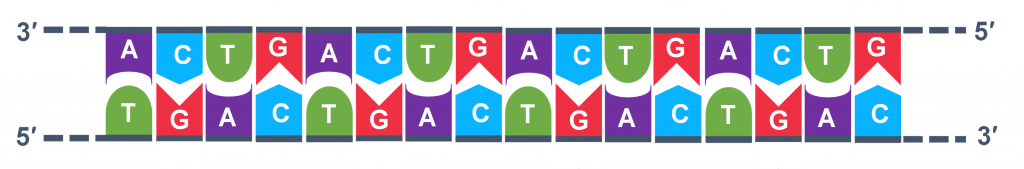
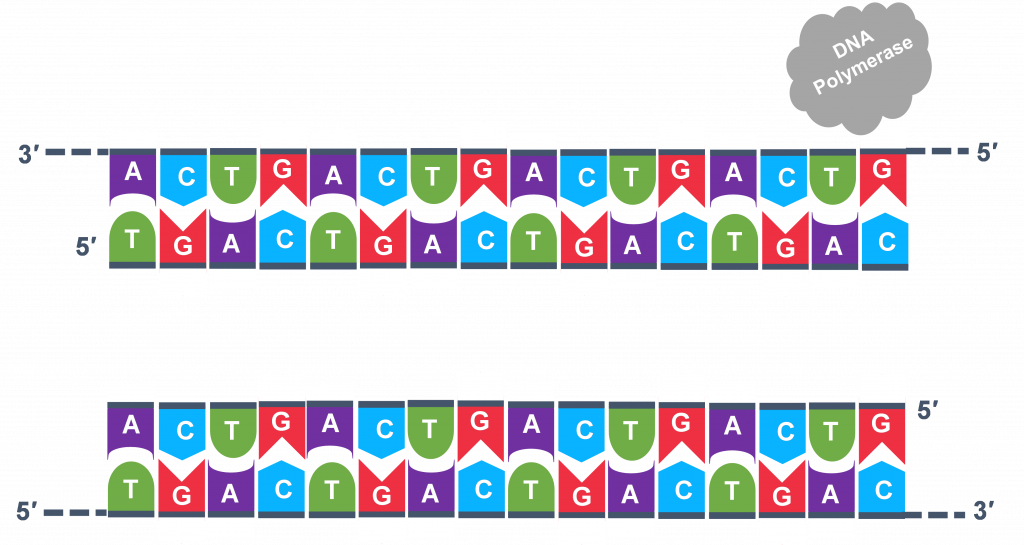
In full general, a single PCR run will undergo 25-35 cycles. The start step for a unmarried wheel is the denaturation step, in which the double-stranded Dna template molecule (Effigy 2) is made single-stranded (Figure 3) . The temperature for this step is typically in the range of 95-100°C, near boiling. The high heat breaks the hydrogen bonds between the strands but does not suspension the carbohydrate-phosphate bonds that concur the nucleotides of a unmarried strand together ( Figure 3 ).
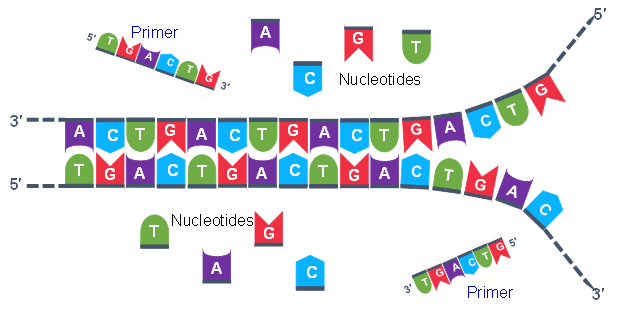
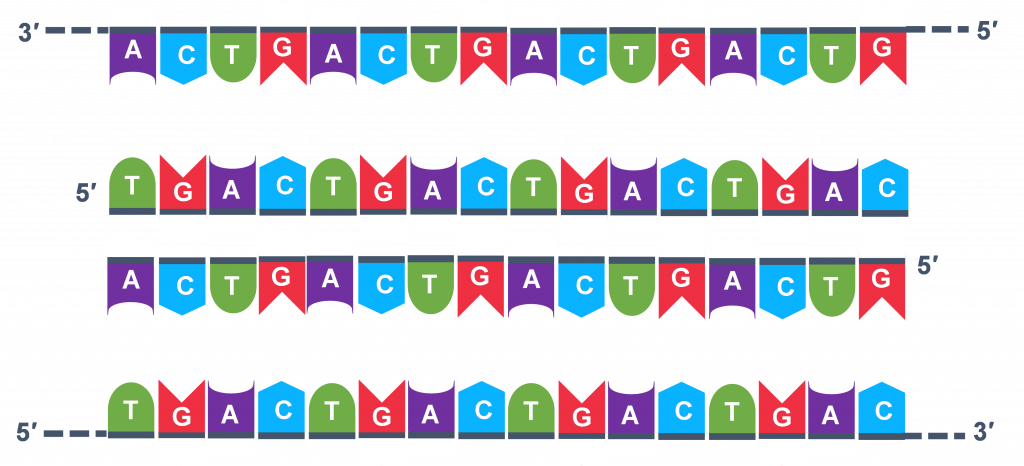
Thousands of copies of the single stranded primers and the individual nucleotides were added to the test tube prior to beginning the cycles. Both the primers and nucleotides will get office of the new Deoxyribonucleic acid strands. The second step in the PCR reaction is to cool the temperature in the test tube to 45-55°C. This is the primer annealing step in which the primers bind to complementary sequences in the single-stranded DNA template. The 2 primers are called the forward and the reverse primer and are designed considering their sequences will target the desired segment of the Deoxyribonucleic acid template for replication (Figure 4).
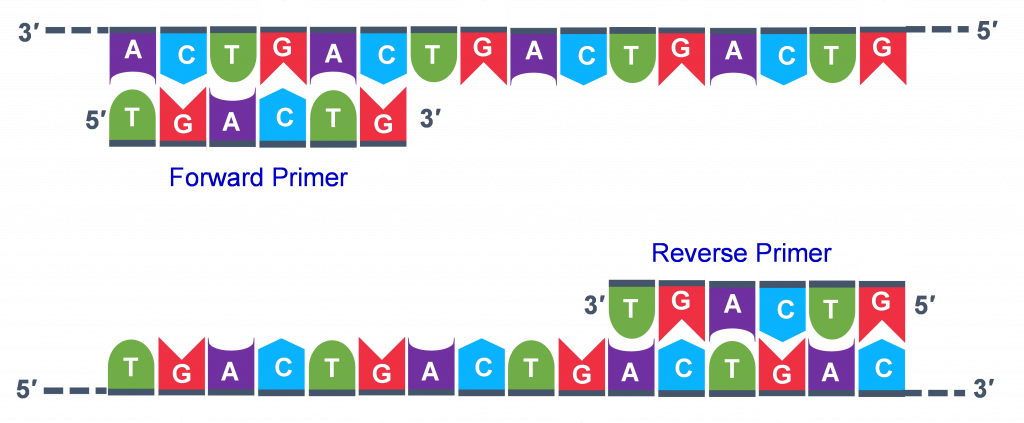
The geneticist planning the PCR analysis must "design" the forrard and reverse primers and then purchase them from a vendor who can synthesize unmarried stranded DNA that has a specific sequence and length. The two almost important criteria for primer design are the following.
- One primer must have a sequence that complements one of the template strands and the other primer must be complementary to the other strand. BOTH strands need to be primed for the replication process.
- The primers must bind and then that their 3′ ends are 'pointing' in the direction of the other primer. This ensures that the sequence between the primers is replicated in the PCR cycles.
Extension: The last PCR step is when the DNA polymerase enzyme reads the template and connects new nucleotides to the primer'south 3' end, extending a new complementary strand of Deoxyribonucleic acid (Figure five). Completion of the final step and the first cycle of PCR, volition make ii double stranded DNA copies from the original template DNA, doubling of the amount of DNA present. The examination tube is heated to around 75°C , optimizing DNA pol. III activity and the newly synthesizing Deoxyribonucleic acid strand is extended every bit the template strand is read by DNA pol. III . The Extension step will run for a few minutes and this footstep complet es 1 PCR cycle.
For bike 2, the denaturation, annealing, and extension steps are repeated (Effigy 6 a, b, c). This time, though in that location will be twice as many Deoxyribonucleic acid template molecules compared to what there was at the starting time of cycle i. Copies are existence made by reading the original template and copies are fabricated by reading the copies made in the previous bike. Therefore, if everything is working correctly, the DNA replication in the test tube is a chain reaction where at the cease of a cycle, at that place is double the amount of that Deoxyribonucleic acid sequence every bit what was found at the kickoff of the cycle.
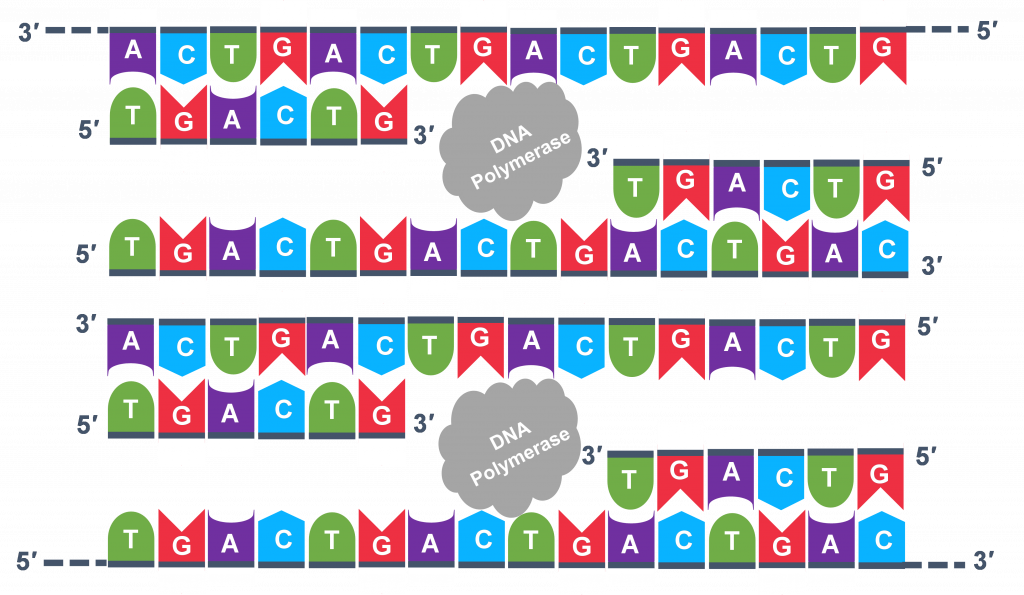
Because thousands of copies of the forward and reverse primer are added at the get-go of PCR, all the unmarried strand templates, both the original, the copies in cycle three and beyond, and the copies of the copies made from previous cycles volition be primed for the extension pace of the cycles.
Thermal cycler

When PCR was first invented, scientists used water baths set at unlike temperatures and the hand transfer of examination tubes at timed intervals to run the PCR reaction. Once the technique became a proven engineering for Dna assay, engineers went to work to create PCR machines. The instrument which heats and cools the DNA samples is chosen a thermal cycler (Figure 7). Each pocket-size tube or sample well in a plate contains all the chemical components needed for a PCR reaction. Adding a specific sample to the reaction mix provides the template DNA. A thermal cycler can be programmed for specific temperatures and the amount of fourth dimension spent at each temperature. The engineered design of thermal cyclers to maximize the accurate replication of the targeted DNA in a small sample volume with the minimum corporeality of time tin exist critical in many applications of PCR.
Taq DNA polymerase
When Dr. Kerry Mullis ran the first PCR experiments, he needed to add together a new sample of Deoxyribonucleic acid politician Three later on each denaturation step. This was because the high temperature needed to denature the double stranded Deoxyribonucleic acid template as well denatured the Dna pol 3 protein construction. The Deoxyribonucleic acid pol III enzyme commonly bachelor to molecular geneticists was from E. coli bacteria and this enzyme had no stability at near boiling temperatures. Fortunately, biologists had been investigating Thermus aquaticus, (Taq) a thermophilic eubacterium found in hot springs (Chien et al. 1976). The Taq version of Dna pol 3 does not hands denature in the hot temperatures required in PCR; plus, it has a skillful efficiency, able to add 60 base pairs/sec at 70°C. Like all other DNA polymerases, Taq Dna political leader III cannot brainstorm DNA replication without the addition of a starting primer. Thus, the discovery of Taq Dna pol Three and the commercial availability of this enzyme fabricated PCR a more reliable and doable applied science which hastened its application to science investigation and diagnostic testing.
Visualizing the Results with Electrophoresis
In one case a PCR reaction has been completed, we need to be able to see the results. To do this, a sample of the PCR mixture is loaded into an agarose gel for electrophoresis. The agarose gel contains a matrix of pores which enables it to dissever DNA fragments based on their sizes. For details about setting upward and running an electrophoresis gel, see Electrophoresis: How Scientist observe fragments of Deoxyribonucleic acid
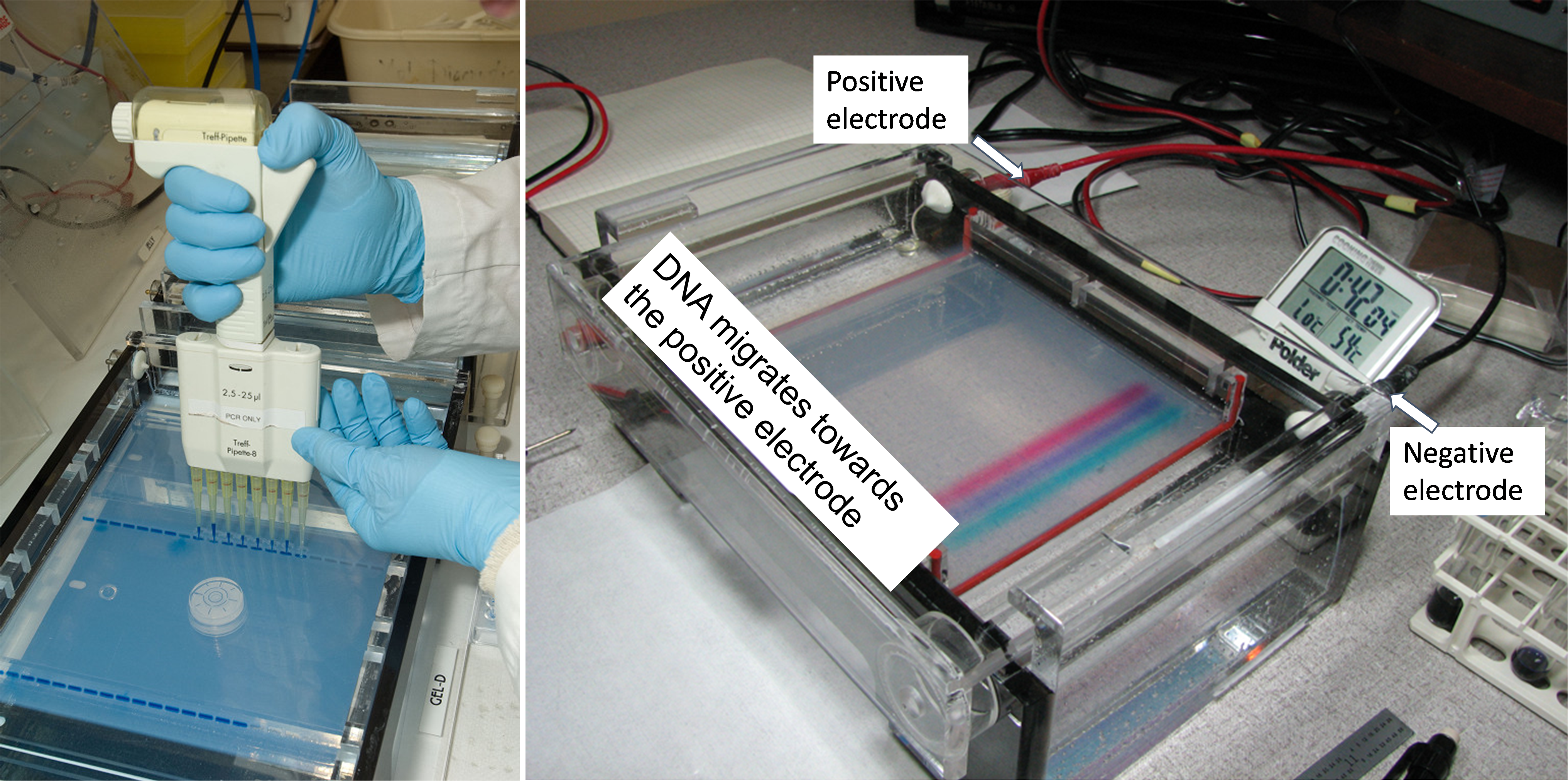
Figure viii shows a picture of a gel electrophoresis gel that is running. The box on the right contains Deoxyribonucleic acid loaded in the agarose gel. The gel placed in an aqueous solution of electrolytes. Depending on the type of dye used, color bands are a dye that was added to the PCR sample before it was loaded into the sample well. This allows for the tracking of the Dna's progression through the gel. Hooked upwards to this gel unit is an electrical power source which provides the strength to motion the Deoxyribonucleic acid through the gel. Since DNA molecules are negatively charged, they volition drift towards the red, positive electrode. Shorter Dna fragments move faster than longer fragments through the pores in the gel.
After the gel is run, the Dna is stained with a chemical that binds specifically to Deoxyribonucleic acid molecules and so will either reflect a specific colour of visible calorie-free or fluoresce a specific color when viewed with ultraviolet light. A single 'band' contains 1000s of individual Deoxyribonucleic acid fragments, all of the same length. Figure 10 illustrates the visual data that can exist obtained from an electrophoresis gel after information technology has run. The electric current uniformly moves all the DNA fragments through a gel in the aforementioned direction. The sample wells at the pinnacle of the gel paradigm thus establish lanes for the DNA samples to move.
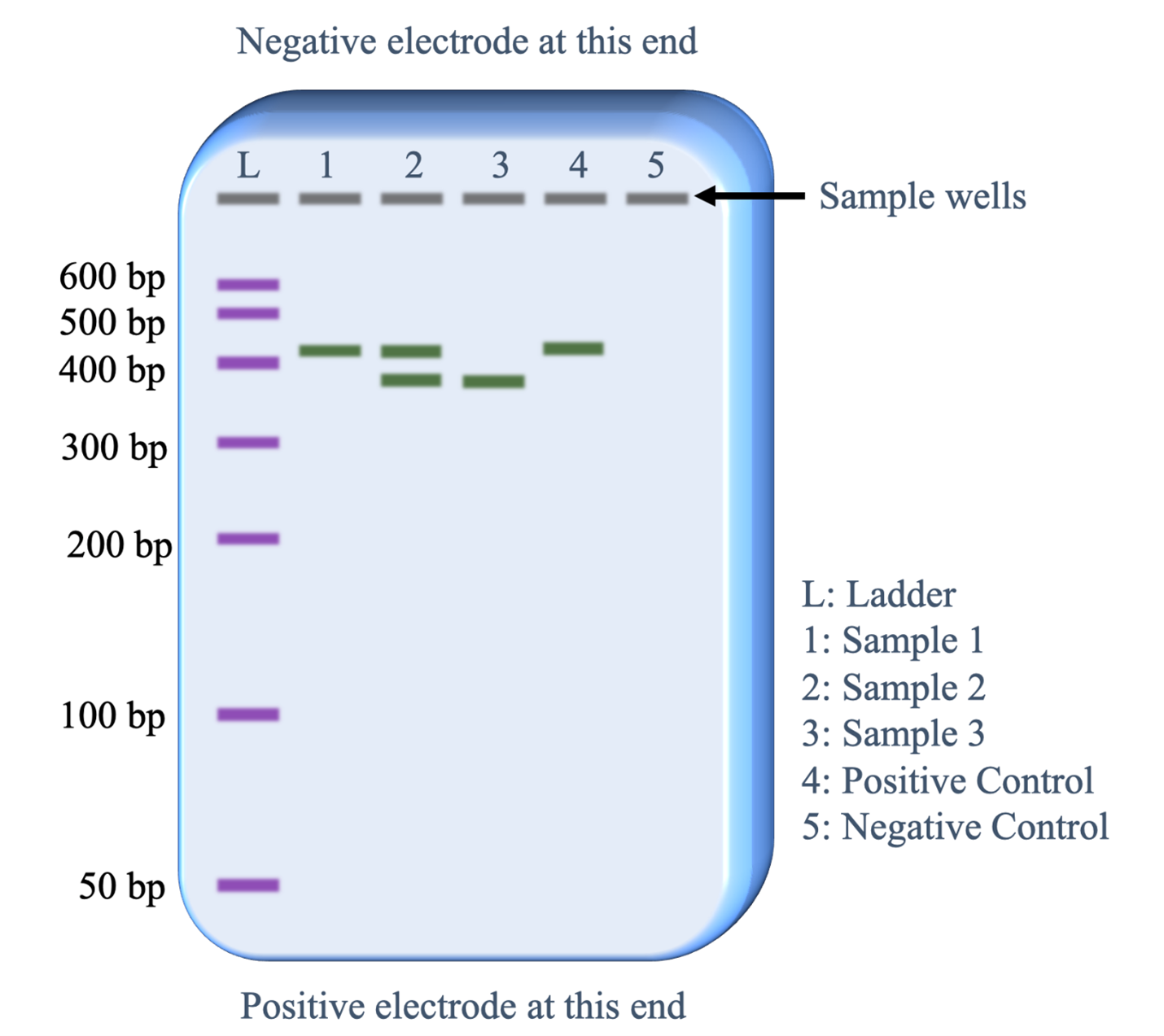
Beneath is a description of what information is revealed from each lane.
- Lane 50: This was loaded with the DNA size ladder that contains copies of vii different lengths of DNA fragments. Commercial vendors of the DNA size ladder provide information on the lengths of each fragment in base of operations pairs (bp). Running this lane provides an estimate of the Deoxyribonucleic acid fragment lengths in the sample lanes (1-5).
- Lane 1: The PCR sample loaded in this lane has copies of a single length of DNA. The length is slightly more than 400 base pairs.
- Lane ii: The PCR sample loaded in this lane has copies of two lengths of DNA. Ane fragment is the aforementioned length as the fragments in lane 1 and the 2nd fragment is slightly less than 400 bp.
- Lane 3: The PCR sample loaded in this lane has copies of one fragment that is the same length as the shorter fragments in lane 2.
- Lane four: The positive control worked as predicted. The sample was set with all the same reagents as the other PCR samples plus a DNA template that was known to contain the sequence targeted past the PCR primers that would generate a 410 base pair fragment.
- Lane 5: The negative control worked as predicted. The sample was set up with all the same reagents every bit the other PCR samples except no template Dna was added. Therefore, nosotros would not expect the PCR reaction to piece of work and the absence of a band of DNA fragments is every bit expected.
Considering the positive and negative controls worked as expected, the biologist can exist confident that the bands of DNA observed in lanes 1,2 and 3 reveal genetic data virtually the individual providing that DNA sample.
Advantages of PCR
PCR speedily became the method of choice for many types of Deoxyribonucleic acid analysis because of several advantages over other DNA detection methods. The first; it is a uncomplicated procedure to set up and run. Fewer steps save time in getting a DNA analysis result. The second is the sensitivity. A very minor amount of template Deoxyribonucleic acid in the sample tin can exist detected. Even merely a few skin cells from ane human pilus incorporate enough DNA, making PCR useful in forensics. The third is that it tin exist designed to accurately differentiate genetic samples that are unlike by equally trivial as single nucleotide in the targeted sequence. Finally, in application where big numbers of samples demand to be subjected to the aforementioned PCR assay, automation and robotic assistance allow the processing of many samples in a very short time. For case, an automated PCR would be critical for testing large numbers of people in hours during a pandemic.
Limitations of PCR
There are some drawbacks of using PCR that i should be aware of too. First, the sequence of the gene or chromosome region existence targeted is required. This limitation is rapidly diminishing as factor and genome sequencing technology and sharing of this sequence through Cyberspace data bases has emerged every bit the norm in genetic analysis. Because of the size of the genomes of living things and a high conservation of gene sequences in many organisms, primers designed for a PCR test must be empirically tested with the proper controls. Biologists tin easily generate simulated positive DNA from PCR that is caused by contamination or lack of specificity in primer design. At that place may likewise be a demand to optimize concentrations of each chemical component. For instance, irresolute the corporeality of Dna template, MgCl2 and Taq polymerase tin can affect both the quantity and quality of bands produced. Some studies take shown that even the make of Taq polymerase tin affect results (Holden et al. 2003). Besides, the temperature cycles may demand to be fine-tuned for a specific PCR test. Finally, as an in vitro Deoxyribonucleic acid replication method, PCR cannot replicate entire chromosomes.
Summary
To briefly highlight the topics of this lesson, remember that PCR is a relatively easy lab technique which amplifies the amount of Dna present, much like living cells do in the beginning stages of a prison cell cycle. There are v chemical components of a PCR reaction: a Deoxyribonucleic acid template, a Deoxyribonucleic acid polymerase, primers, nucleotides, and a buffer. The three temperature steps for 1 cycle are the denaturation, primer annealing and extension steps. There are now many variations and uses of PCR ranging from forensics to genomic studies to identifying transgenic crops.
Lesson Activities
Picket these videos to acquire more about PCR:
- Polymerase Concatenation Reaction (PCR) from DNA Learning Center
- Polymerase Chain Reaction (from UNL)
Source: https://bio.libretexts.org/Bookshelves/Genetics/Genetics,_Agriculture,_and_Biotechnology_%28Suza_and_Lee%29/01:_Chapters/1.04:_PCR_and_Gel_Electrophoresis
Posted by: buckleysurnoted1978.blogspot.com


0 Response to "What Is The Template Strand From The Gel"
Post a Comment